Research
Enable data collection

Most crystallography beamlines are optimized for collecting bright diffraction signals from cryogenically frozen samples to simplify transport and reduce radiation damage. To study protein dynamics with the much weaker and complex diffuse scattering signals, we need unfrozen samples, which introduces new challenges. We are developing practical strategies for measuring diffuse scattering at room temperature, addressing experimental constraints to make this accessible for non-specialists.
Make software accessible
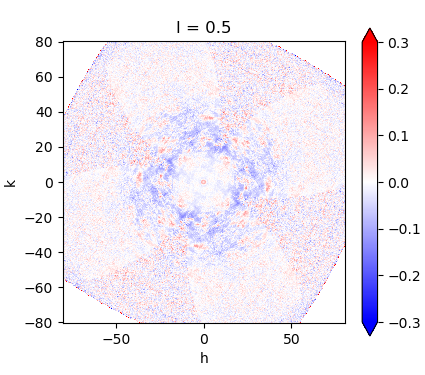
Until now, diffuse scattering analysis has been carried out by a small number of experts using custom-built software. We will build on the state of the art to develop tools that are intuitive, robust, and usable by the broader community.
Expand public datasets
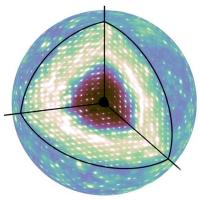
With only a handful of publicly available diffuse scattering datasets today, we will lead an effort to populate a public database with compelling, high-quality examples, expanding the resources available to the community and lowering barriers for others to analyze, interpret, and apply diffuse scattering data in their research.
Improve models of protein dynamics
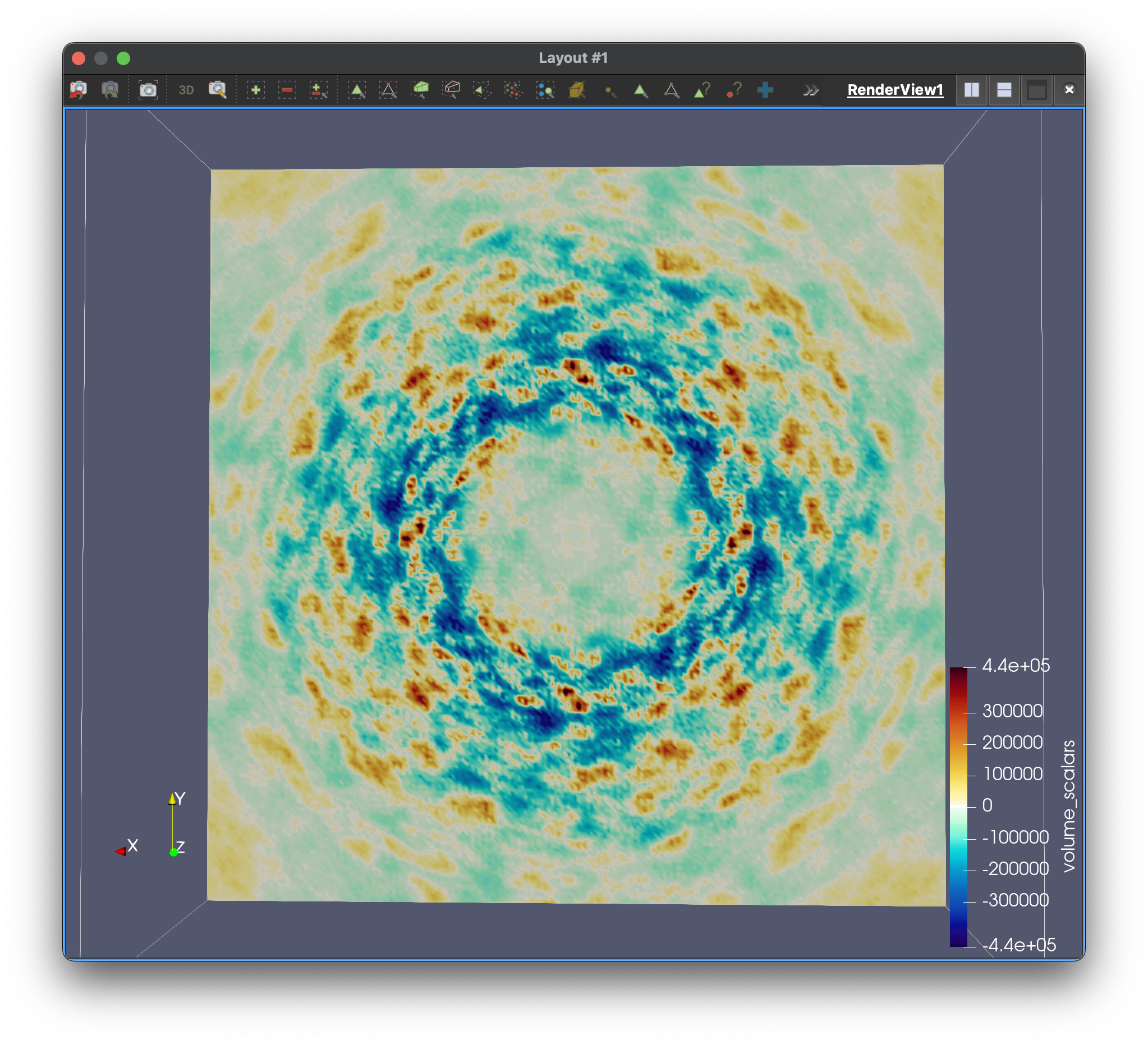
To extract meaningful biological insights from diffuse scattering data, accurate modeling is essential. At present, we model protein motions separately from Bragg peaks and from diffuse scattering, then compare them, but the results often disagree. We are developing algorithms that learn directly from experimental data, integrating information from both Bragg and diffuse scattering to develop a unified, consistent model.
Encoding dynamic models
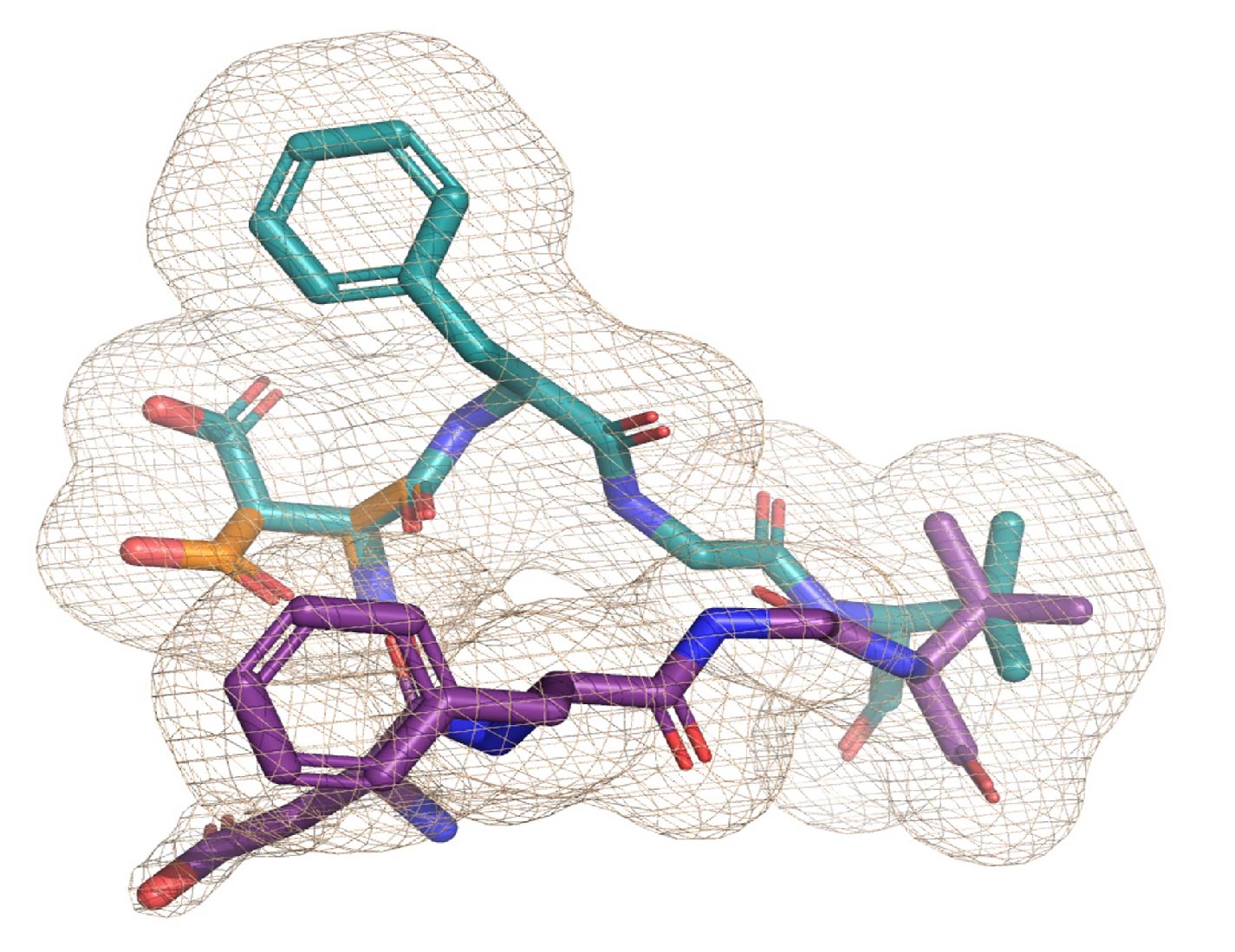
Correctly representing and encoding the outputs of structural modeling is essential for translating them into biological insight. To enable AI co-driven scientific discovery, these representations must be both machine-readable and human-interpretable to leverage the full richness of existing and future experimental data and provide interpretable feedback to guide human insight. We are developing infrastructure to improve model encoding and data architecture, allowing the others to learn from the rich experimental data collected in this project.