Dynamic Structural Biology
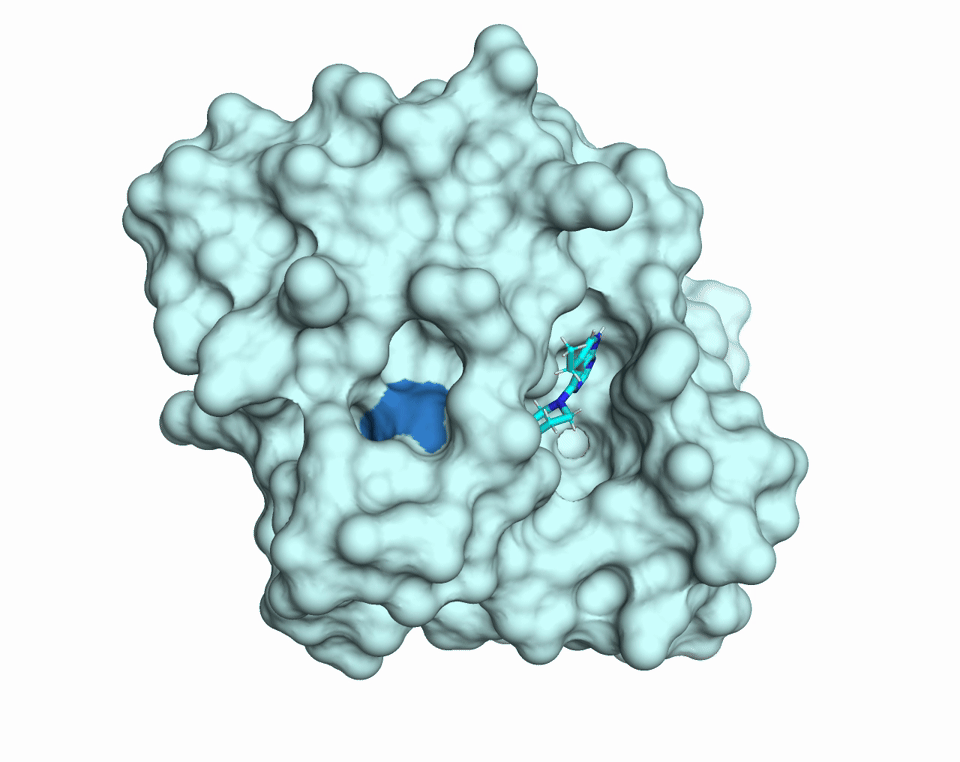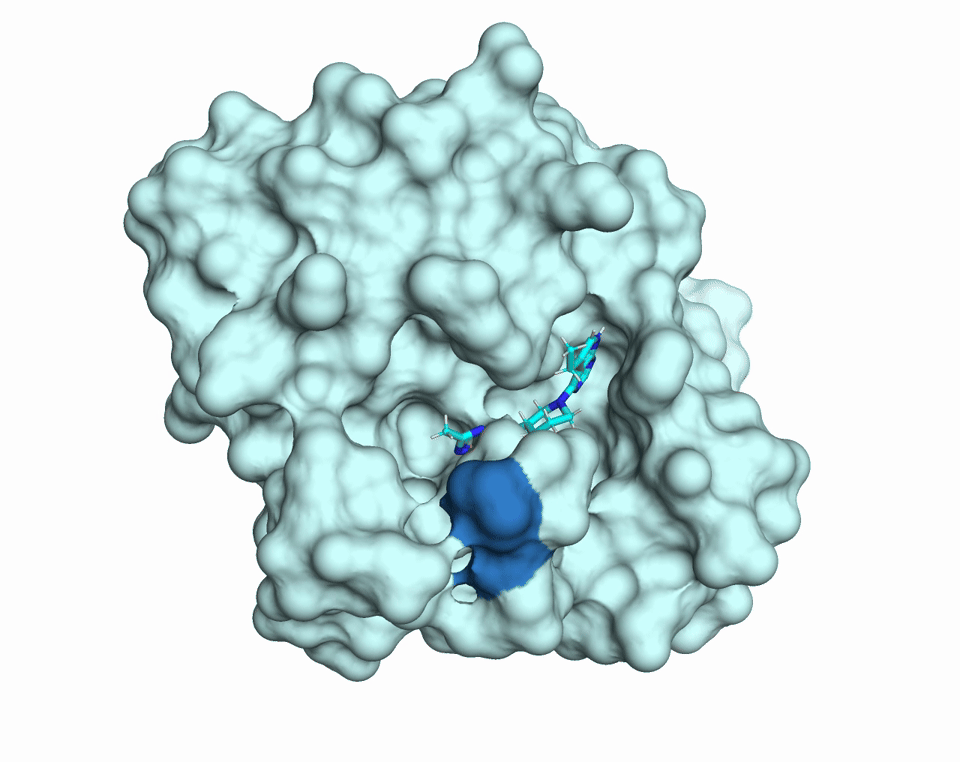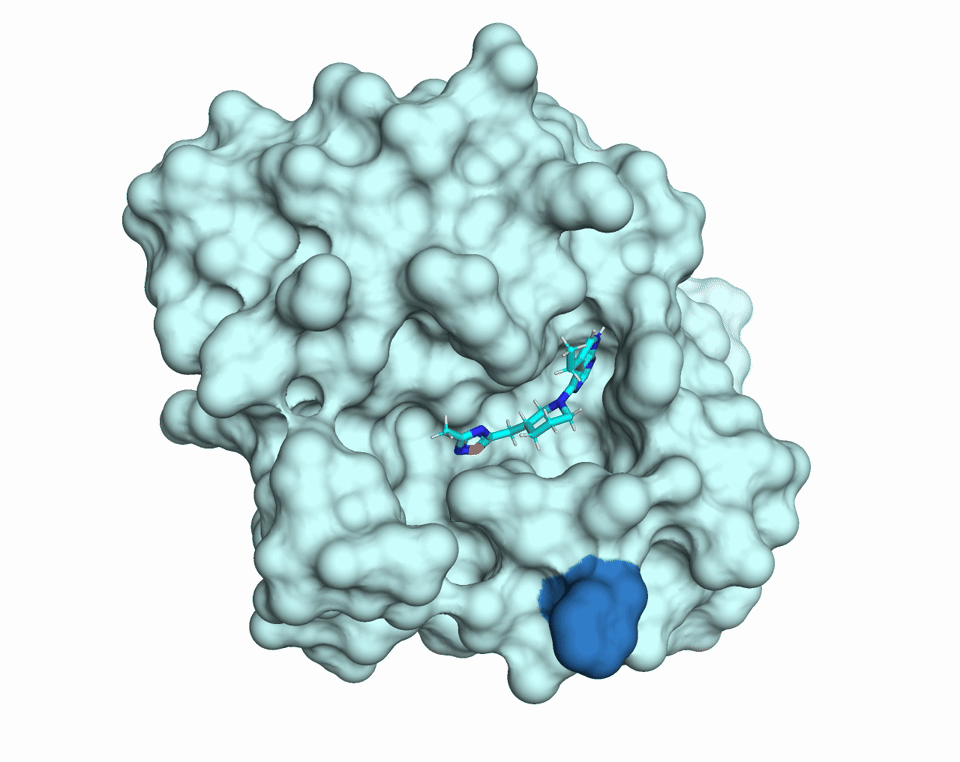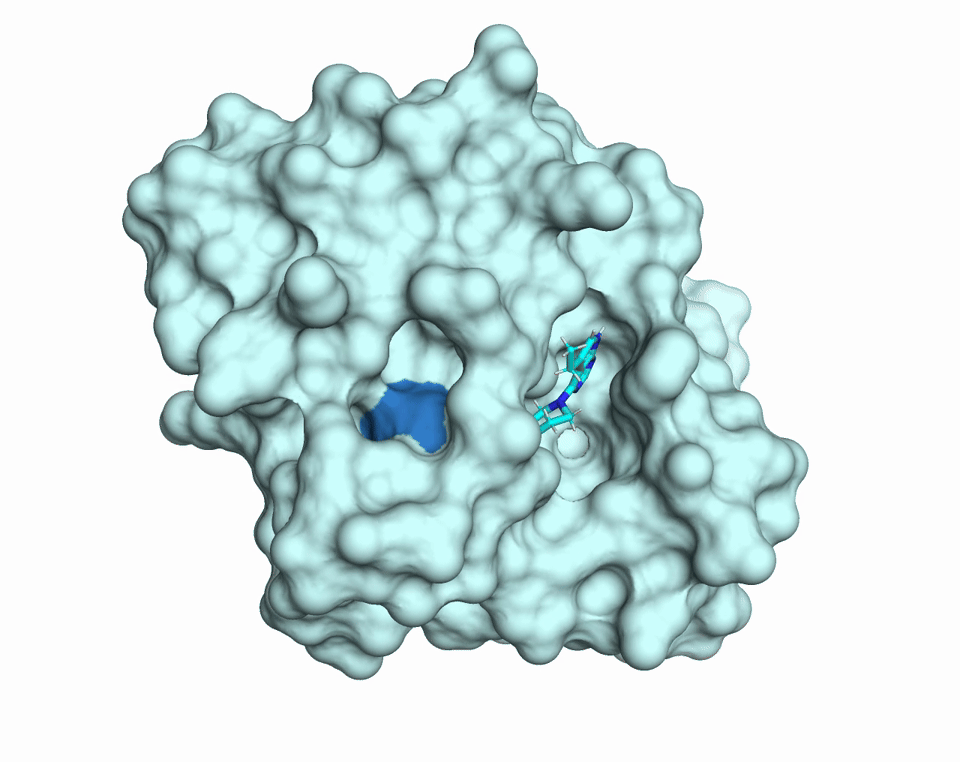
Proteins are dynamic molecules. They move. They breathe. This movement enables their function. The diffUSE Project is building the methods, tools, and infrastructure to make protein motion visible, measurable, and usable — transforming how we discover drugs, design biology, and understand disease.
Reimagining How We Approach Science
We believe that unlocking protein dynamics requires not just new methods but also new ways we work together as scientists. By building openly, we aim to enable the collection of dynamic data at the scale and diversity needed to uncover the fundamental principles of biology.
The DiffUSE Hub
Our open science repository for unlocking protein dynamics, under active development. Notebook
The DiffUSE Logbook
An open science logbook for cataloging X-ray datasets, synchrotron notes, and unfiltered analyses. Blog
DiffUSE Dispatches
Blog posts from our scientists on methods, discoveries, and open science.
Latest Posts from our Scientists
Shake It Up!
MD simulations of changes in diffuse scattering depending on ligand binding
DiffUSE January 2026 Retreat: From Coast to Coast, Diffuse Scattering Reproduces
A report on our January 2026 all-hands meeting
In the Cloud
Next steps in diffUSE MD simulations